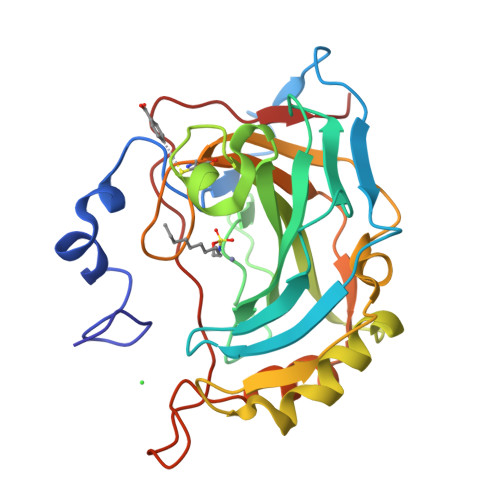
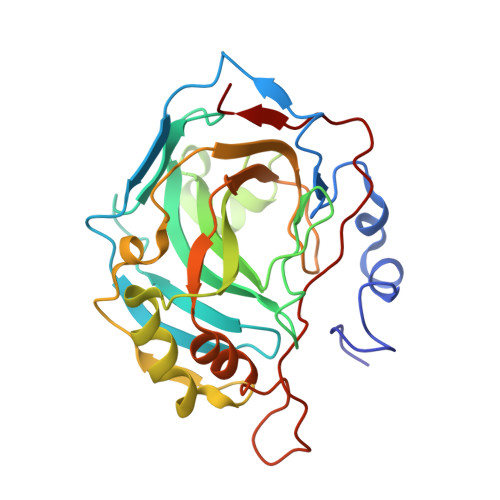
Carbonic anhydrase inhibitors. Comparison of aliphatic sulfamate/bis-sulfamate adducts with isozymes II and IX as a platform for designing tight-binding, more isoform-selective inhibitors
Vitale, R.M., Alterio, V., Innocenti, A., Winum, J.-Y., Monti, S.M., De Simone, G., Supuran, C.T.(2009) J Med Chem 52: 5990-5998
- PubMed: 19731956
- DOI: https://doi.org/10.1021/jm900641r
- Primary Citation of Related Structures:
3IBI, 3IBL, 3IBN, 3IBU - PubMed Abstract:
Two approaches were used to design inhibitors of the metalloenzyme carbonic anhydrase (CA, EC 4.2.1.1): the tail and the ring approaches. Aliphatic sulfamates constitute a class of CA inhibitors (CAIs) that cannot be classified in either one of these categories. We report here the detailed inhibition profile of four such compounds against isoforms CAs I-XIV, the first crystallographic structures of these compounds in adduct with isoform II, and molecular modeling studies for their interaction with hCA IX. Aliphatic monosulfamates/bis-sulfamates were nanomolar inhibitors of hCAs II, IX, and XII, unlike aromatic/heterocyclic sulfonamides that promiscuously inhibit most CA isozymes with low nanomolar affinity. The bis-sulfamates incorporating 8 or 10 carbon atoms showed higher affinity for the tumor-associated hCA IX compared to hCA II, whereas the opposite was true for the monosulfamates. The explanation for their interaction with CA active site furnishes insights for obtaining compounds with increased affinity/selectivity for various isozymes.
Organizational Affiliation:
Istituto di Chimica Biomolecolare-CNR, Pozzuoli, Italy.